We have screened >300 antibodies as part of this project, generating a massive amount of data and revealing important problems with histone PTM antibodies. However, some of these problems may not be apparent from searching for a single PTM or an individual antibody. Thus, we have crafted this section to guide scientists through the website, as well as highlight the major findings of this study.
- Why are the antibodies defined by an E number? What is this?
- Luminex® and SNAP-ChIP® Spike-in data : What is the difference?
- Why are different lots of the same antibody tested?
- Does it matter if my antibody is polyclonal or monoclonal?
- What does antibody efficiency mean? Is it as important as specificity?
- My antibody has been cited hundreds of times, but fails SNAP-ChIP; is this possible?
- Once I have found a SNAP-ChIP validated antibody, do I still need to use spike-in controls?
Additional questions are answered on our Frequently Asked Questions (FAQ) page.
Why are the antibodies defined by an E number? What is this?
The E number is a unique identifier we gave each antibody to enable unbiased testing, and corresponds to the labels we used in our recent paper (insert ref to new paper HERE). If you hover over the E number the company, catalog number, and lot number appear, making it easy to look up your favorite antibody for purchase on an external site.
If you are navigating to our website from the recent manuscript and are looking for information by the E number- not a problem! Simply type the E number into the search bar.
For some antibodies the E numbers are the same but have a different integer after the decimal. For example, there are antibodies to H3K36me1 that are labelled E0050.1 and E0050.2. This indicates that the antibodies are different lots of the same antibody (more on lot-testing below).
Luminex and SNAP-ChIP® Spike-in data : What is the Difference?
Upon searching for your favorite histone modification, you will be able to toggle between heatmaps displaying results from SNAP-ChIP and Luminex antibody screening. A brief description is given below; for more on how these experiments were performed, see our METHODOLOGY page.
“Luminex” refers to our first-pass method for rapidly screening hundreds of antibodies to narrow down which antibodies should be validated using SNAP-ChIP Spike-ins. This triage assay uses nucleosomes bound to uniquely barcoded Luminex xMAP® beads, and antibody binding is detected on the FLEXMAP 3D®. “Passing” the Luminex test requires that an antibody exhibited <10% cross-reactivity to off-target histone PTMs. The Luminex heatmaps include data for all antibodies examined by the assay, and are sorted such that the best performing antibodies are at the top of the heatmap. Antibodies that failed Luminex testing are also included in these heatmaps.
“SNAP-ChIP” denotes our provides final antibody validation test, using SNAP-ChIP Spike-in Panels to characterize antibody performance in a ChIP experiment. SNAP-ChIP Spike-in testing is distinct from Luminex in that it:
- Validates the antibody against a defined chromatin substrate in an actual ChIP workflow.
- Provides information on antibody efficiency (i.e. enrichment of on-target PTM relative to Input).
Antibodies that passed SNAP-ChIP Spike-in testing, and thus are certified for ChIP and ChIP-seq experiments, displayed >5% efficiency and <20% off-target binding. We have included data for all antibodies tested in SNAP-ChIP, including those that failed (see more on this point in the next section). The best performing antibodies are at the top of the heatmap.
We do not recommend using antibodies that fail Luminex and/or SNAP-ChIP criteria for ChIP-seq experiments, as the data will be contaminated by a significant amount of off-target signal, which could alter biological analyses and conclusions.
Why are some antibodies only displaying partial results in SNAP-ChIP Spike-in testing heatmaps?
Prior to development of our Luminex triage strategy, we performed preliminary antibody testing using a scaled-down SNAP-ChIP Spike-in panel. In these “Phase I” SNAP-ChIP Spike-in assays, antibodies were profiled against a subset of SNAP-ChIP controls, representing PTMs most closely related to the on-target PTM.
If the antibody failed to meet our standard SNAP-ChIP criteria against this smaller PTM panel (i.e. <20% off-target binding, >5% enrichment), then the antibody was not tested using the full spike-in panel. In addition, if that particular lot of antibody was no longer available, we may not have tested it in a full SNAP-ChIP Spike-in experiment. However, we still opted to include these data in our heatmaps, as some of these antibodies may be used by researchers in the field.
Luminex vs. SNAP-ChIP Spike-in results : Which data should I use?
If you are simply looking for the best antibody to a given histone PTM, toggle to the SNAP-ChIP data and look at antibodies towards the top of the heatmap. The antibodies are ranked such that those with the greatest specificity and efficiency are listed at the top of the heatmap. Click on the unique identifier (the E #) to bring up all data for that specific antibody, including any Luminex data and information on other tested lots (see more on lot-testing below).
It is also useful to look at the heatmaps to observe, in general, how well commercial antibodies perform for a given histone PTM target. Some PTMs have more specific antibodies than others. We had a particularly difficult time finding good antibodies for H3K9me2, H3K36me2, and H3K27ac, as many antibodies to these targets failed Luminex and/or SNAP-ChIP preliminary testing. However, despite these challenges, we were able to identify highly specific antibodies to most of the PTMs in our panels.
Despite the fact that most antibodies failed SNAP-ChIP Spike-in testing, the majority of these antibodies are described as “ChIP-grade.” This is likely a result of validating antibodies with modified histone peptide arrays, whose linear epitopes fail to model the complex chromatin structures seen in vivo. These results highlight the necessity of nucleosome-based, in-application testing to accurately determine antibody performance in ChIP.
Why are different lots of the same antibody tested?
At the bottom of each antibody page, we have included a table of pass / fail testing results for other antibodies with the same catalog number and vendor, but a different lot number. We began sourcing multiple lots of the same antibody, where available, in the later stages of our study once it became apparent that lot testing is essential for ChIP antibody validation.
This is best demonstrated by using a representative example. Figure 1 shows SNAP-ChIP Spike-in data for two lots of the same H3K27me1 antibody. The lot shown in Figure 1A (E0028.3) passed our SNAP-ChIP Phase I testing, and also passed final SNAP-ChIP Spike-in testing against our entire histone lysine methylation panel (see antibody page for full testing data). The antibody displayed high efficiency, with >25% enrichment of target vs. input.
Notably, an antibody from the same company / catalog number, but from a different lot (E0028.5) exhibited strikingly different specificity and efficiency against SNAP-ChIP Spike-ins (Figure 1B). E0028.5 has high binding to both unmodified nucleosomes and related PTMs H3K27me2 and me3. In addition, this lot also displays poor enrichment (4.8%). This lot of antibody did not pass SNAP-ChIP testing and is not recommended for ChIP-seq studies.
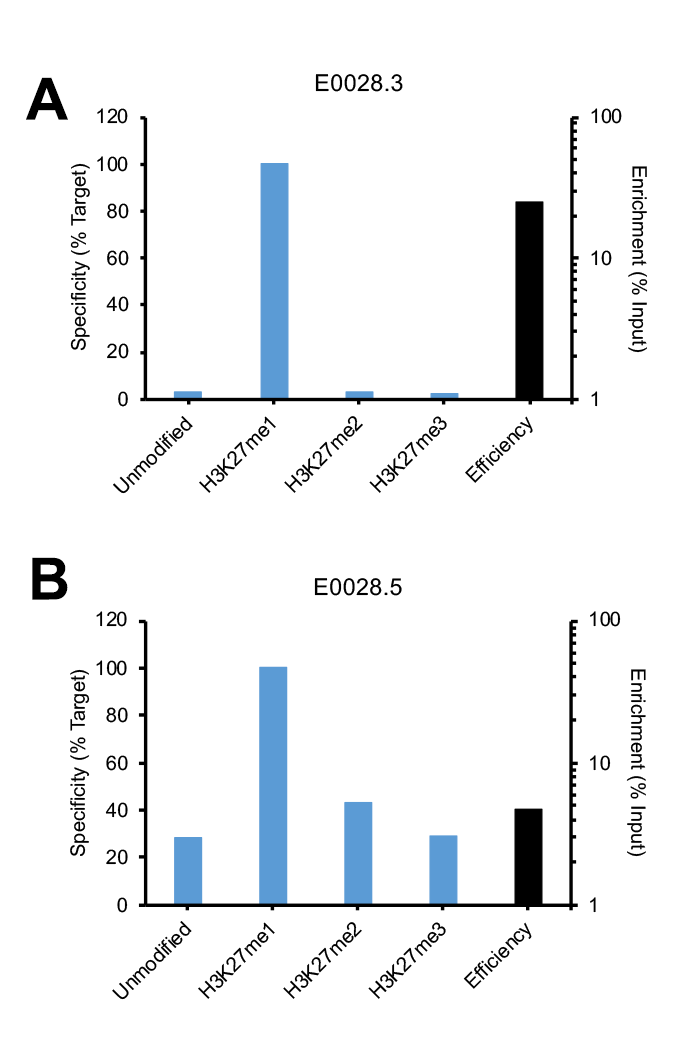
Figure 1: SNAP-ChIP Spike-in testing of different lots of the same polyclonal H3K27me1 antibody (E0028). (A) Lot 3 met SNAP-ChIP testing metrics, with <20% cross reactivity >5% enrichment of H3K27me1 vs. input chromatin, while (B) Lot 5 failed to meet these same metrics.
It isn’t altogether surprising that we observed lot-to-lot changes in antibody specificity and efficiency, as batch variation effects are known to impact antibody binding1, 2. However, in the context of histone PTMs it has not been fully documented, and typically scientists select antibodies to these targets based on citations or past performance, regardless of lot number.
Although our early data on lot variation is somewhat variable, these information will increase in the future as we source additional antibodies. In the meantime, we strongly recommend lot-testing your antibodies prior to using them for any ChIP-seq assays.
Does it matter if my antibody is polyclonal or monoclonal?
The predominant narrative in the field is that monoclonal antibodies are always preferred over polyclonal antibodies. However, in our screening, we found that many polyclonal AND monoclonal antibodies failed to pass SNAP-ChIP testing, and both exhibited lot variability.
Figure 2 provides a key example of a monoclonal H3K27me3 antibody that displays lot variation in specificity. Lot 1 (Figure 2A) passes SNAP-ChIP Spike-in testing, with <20% off-target binding and a strong efficiency of >10%. Lot 3, shown in Figure 2B, still displays moderate efficiency of approximately 10%, but has increased off-target binding for unrelated lysine methyl marks, including H3K4me1, H3K36me1, and H4K20me1. Many monoclonal and polyclonal antibodies display variation in efficiency from lot-to-lot (see Figure 1; for an example of monoclonal antibody displaying changes in efficiency, see H4K20me1 antibody E0117.2 vs E0117.3).
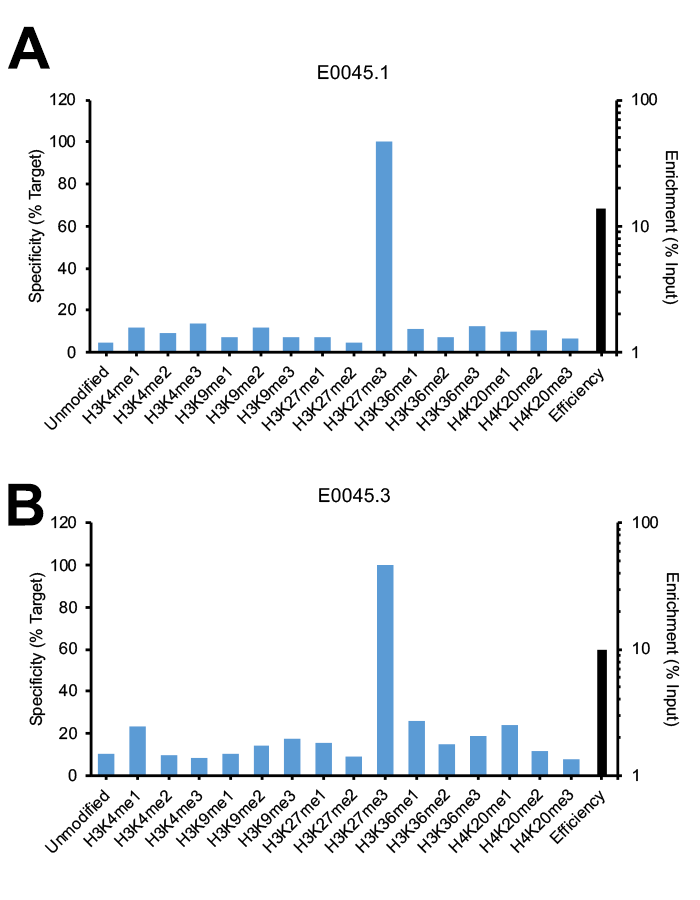
Figure 2: SNAP-ChIP Spike-in testing of different lots of the same monoclonal H3K27me3 antibody (E0045). (A) Lot 1 met SNAP-ChIP testing metrics, with <20% cross reactivity >5% enrichment of H3K27me3 vs. input chromatin, while (B) Lot 3 failed to meet these same metrics.
Just because your antibody is monoclonal doesn’t mean it will pass SNAP-ChIP Spike-in testing, or that it will be consistent across production lots. Every new lot of antibody you purchase should be tested against a physiological chromatin control in the context of a ChIP experiment, such as SNAP-ChIP Control Panels.
What does antibody efficiency mean? Is it as important as specificity?
The efficiency of an antibody in ChIP refers to the enrichment of the target following immunoprecipitation (IP). Typically, enrichment is determined by performing qPCR for a positive control locus, calculating the signal from the ChIP reaction as a percent of the signal from input chromatin. However, if the relative abundance of the epitope in bulk chromatin is unknown, this information may not be very useful, or accurate.
SNAP-ChIP Spike-in validation provides a quantitative metric for efficiency, by calculating the recovery of on-target SNAP-ChIP nucleosomes that were spiked into bulk chromatin. To pass SNAP-ChIP certification testing, the antibody must recover at least 5% of the spike-in nucleosome (vs. spiked input).
This metric is especially important when profiling histone PTMs with low abundance in the genome, such as methylation on H3K43. In fact, we have found that a high antibody efficiency correlates with increased sample recovery (i.e. more material to produce high quality sequencing libraries) and improved signal : noise (S/N) in ChIP and ChIP-seq (see Figure 3)4.
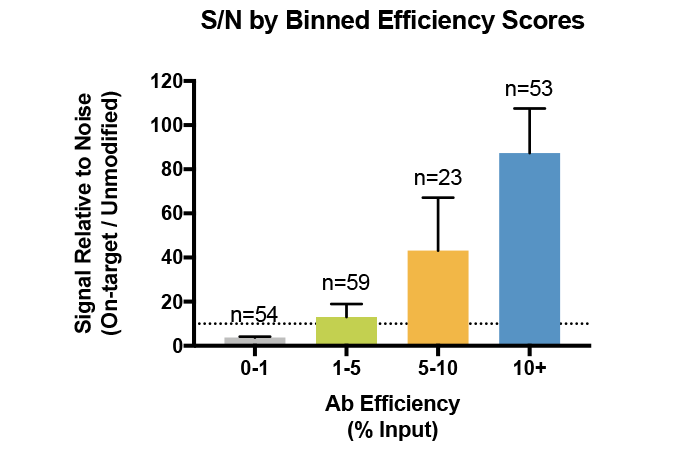
Figure 3: Average signal-to-noise (S/N) for H3K4me0, me1, me2, and me3 antibodies tested against SNAP-ChIP Spike-in Controls. Antibodies are binned by efficiency score, revealing that improved enrichment correlates with improved recovery and S/N. Data sourced from Shah et al. 2018.
Compared to specificity, we give efficiency equal weight in our SNAP-ChIP Spike-in criteria; if an antibody fails to meet either metric, it is not considered SNAP-ChIP certified. The addition of efficiency to our criteria ensures that we identify only the highest quality antibodies for future ChIP-seq studies.
My antibody has been cited hundreds of times, but fails testing against SNAP-ChIP Spike-ins; is this possible?
This was actually a major conclusion of our 2018 study, in which we reported that a highly cited H3K4me3 antibody displayed substantial cross reactivity with H3K4me2 (see Figure 4)4. Importantly, this off-target binding was not detected by standard testing using a modified histone peptide array, again highlighting the importance of using a defined chromatin substrate for in-application antibody testing.
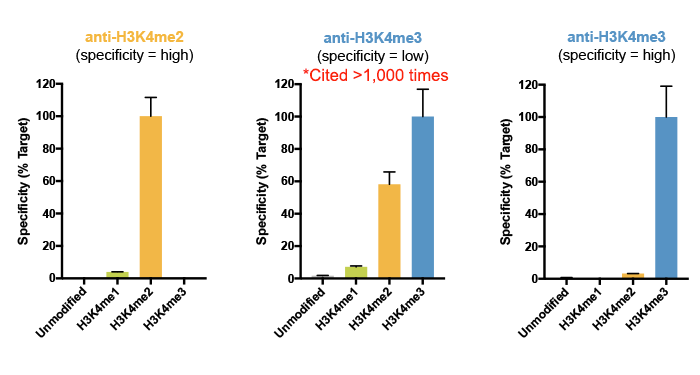
Figure 4: SNAP-ChIP Spike-in results for a highly specific H3K4me2 antibody (left) and H3K4me3 antibody (right). In the center is SNAP-ChIP specificity data for a widely cited H3K4me3 antibody, which exhibits high levels of off-target binding to H3K4me2. Data sourced from Shah et al. 2018.
Our continued work on other classes of histone PTMs (i.e. acylation) has generated similar data. Going forward, we do not recommend relying on previous publications as the primary source for selecting an anti-histone PTM antibody.
Once I have found a SNAP-ChIP validated antibody, do I still need to use spike-in controls?
EpiCypher recommends using SNAP-ChIP Spike-ins in all ChIP-seq experiments, even if you are using a SNAP-ChIP certified antibody. ChIP is an inherently inconsistent method, due to the enrichment of small subsets from large, fragmented pools of excess chromatin. Even when using a highly reliable antibody, we sometimes see differences in SNAP-ChIP Spike-in specificity or enrichment between experimental replicates.
SNAP-ChIP Spike-ins can be an incredibly useful tool in these circumstances, to gauge the success of your ChIP prior to library prep and next generation sequencing (NGS). We typically analyze SNAP-ChIP Spike-in barcodes by qPCR following IP, which confirms specificity and efficiency, and then move ahead with next NGS. Any replicates that fall outside standard SNAP-ChIP testing metrics are not used in sequencing studies.
In addition, SNAP-ChIP Spike-ins can be used to normalize ChIP-seq reactions; for these purposes, we point readers to recent studies, both from EpiCypher and independent groups (here, here, and here)4-8.
REFERENCES
- Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature, 2015. 518(7537). PubMed PMID: 25652980
- Leroy G, et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics Chromatin, 2013. 6(1). PubMed PMID: 23826629
- Shah RN, et al. Examining the Roles of H3K4 Methylation States with Systematically Characterized Antibodies. Mol Cell, 2018. 72(1). PubMed PMID: 30244833
- Lam KG, et al. Cell-type-specific genomics reveals histone modification dynamics in mammalian meiosis. Nat Commun, 2019. 10(1). PubMed PMID: 31444359
- Tay RE, et al. Hdac3 is an epigenetic inhibitor of the cytotoxicity program in CD8 T cells. J Exp Med, 2020. 217(7). PubMed PMID: 32374402
- Grzybowski AT, et al. Calibrating ChIP-Seq with Nucleosomal Internal Standards to Measure Histone Modification Density Genome Wide. Mol Cell, 2015. 58(5). PubMed PMID: 26004229
- Vallianatos CN, et al. Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. bioRxiv, 2020.